
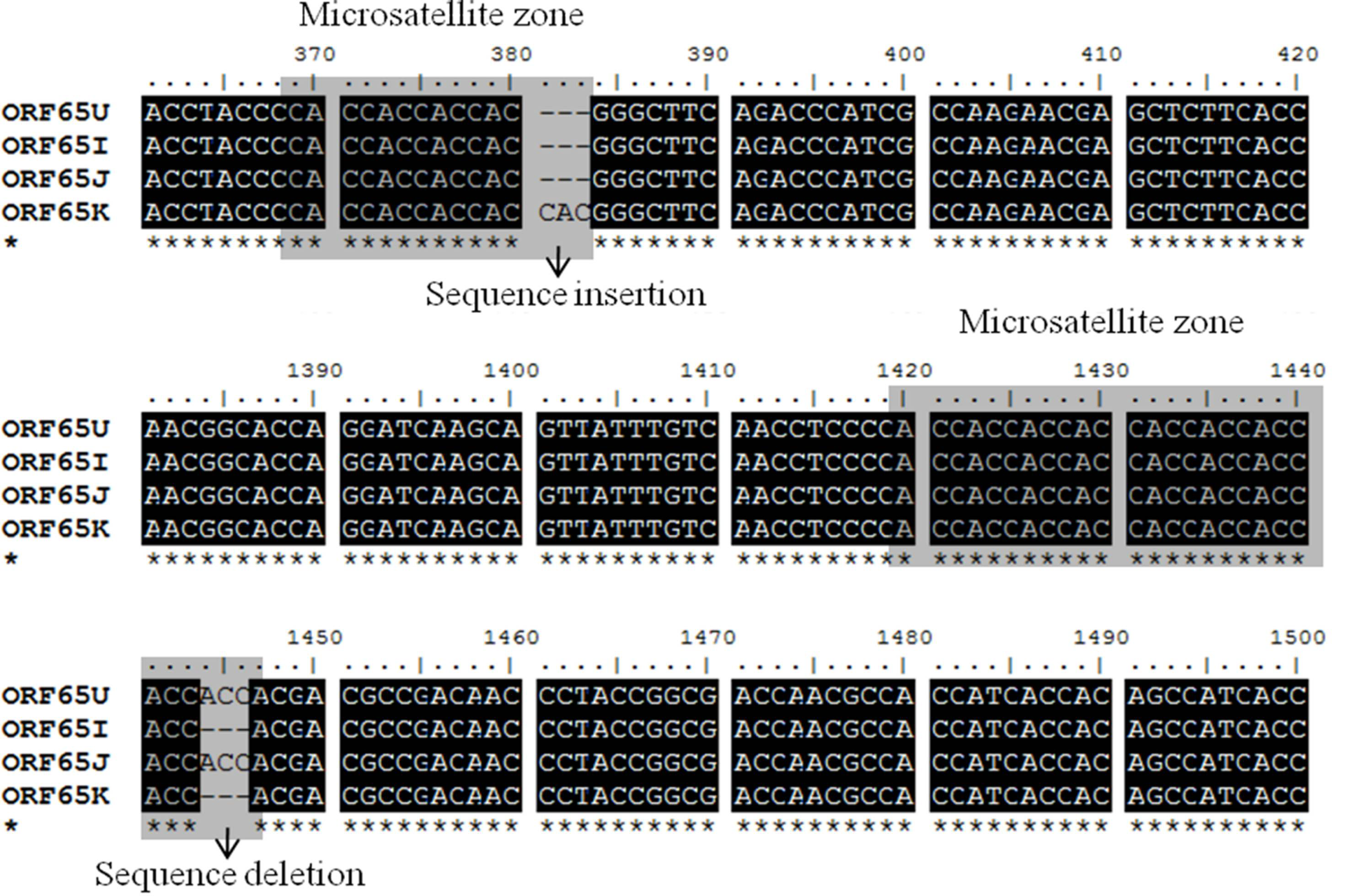
This should be the case anyway if the sequences have been automatically aligned using a program like Clustal. Therefore alignment gaps must be in multiples of 3 (representing an aa deletion/insertion) and at present must be indicated by a '-' symbol.Īlignment must be solely of coding region and be in reading frame 0 to achieve meaningful resultsĪlignment must therefore be a multiple of 3 nucleotides long.Īll sequences must be the same length (including gaps). Use the subroutine "aa_to_dna_aln" in Bio::Align::Utilities to achieve this.
In order to use these methods there are several pre-requisites for the alignment.ĭNA alignment must be based on protein alignment. This method works well so long as the nucleotide frequencies are roughly equal and there is no significant transition/transversion bias. All are implementations of the Nei-Gojobori evolutionary pathway method and use the Jukes-Cantor method of nucleotide substitution. There are also three methods to calculate the ratio of synonymous to non-synonymous mutations. Listed in brackets are the pattern which will match Several different distance method calculations are supported. Work is underway to correct them, but do not expect this code to give you the right answer currently! Use dnadist/distmat in the PHLYIP or EMBOSS packages to calculate the distances. The routines are not well tested and do contain errors at this point. This object contains routines for calculating various statistics and distances for DNA alignments. file => 't/data/nei_gojobori_test.aln') My $in = Bio::AlignIO->new(-format => 'fasta', # and for measurements of synonymous /nonsynonymous substitutions # My $jcmatrix = $stats->distance(-align => $aln,

My $alignin = Bio::AlignIO->new(-format => 'emboss', My $stats = Bio::Align::DNAStatistics->new() Bio::Align::DNAStatistics - Calculate some statistics for a DNA alignment SYNOPSIS use Bio::AlignIO


 0 kommentar(er)
0 kommentar(er)
